Background on Saccharomyces cerevisiae
The term "yeast" derives from words related to "foaming" or "rising," directly associated with the processes of beer and bread production, especially with the alcoholic fermentation of sugary liquids. Generally, when people refer to "yeast" in daily life, they are specifically referring to the brewing yeast (Saccharomyces cerevisiae), which plays a crucial role in the production of alcoholic beverages and bread. However, strictly speaking, "yeast" does not specifically denote a single species, but rather a general term for a group of eukaryotic organisms primarily composed of single cells, which reproduce asexually mainly through budding or fission, namely the genus Saccharomyces.
Currently, the genus Saccharomyces comprises eight natural species: S. cerevisiae, S. paradoxus, S. mikatae, S. kudriavzevii, S. arboricola, S. uvarum, S. eubayanus, and S. jurei, as well as two natural hybrids: S. bayanus and S. pastorianus. Among these yeasts, brewing yeast is the most important and widely used representative in fermentation environments.
Brewing yeast is a type of single-celled ascomycetous fungus, with an optimal growth temperature ranging between 25-30°C. It possesses 16 chromosomes, with a nuclear genome size of 12068 kilobases, containing approximately over 6000 genes, of which around 5570 genes are predicted to be involved in protein expression. Compared to other eukaryotic organisms, brewing yeast has a smaller genome and was the first eukaryote to have its entire genome sequenced. Despite its simple structure, brewing yeast can accurately express nearly all biological functions present in eukaryotes. It possesses detailed genome annotation information, is easy to culture, amenable to genetic editing operations, and facilitates phenotype observation. These biological characteristics have made brewing yeast one of the most powerful eukaryotic models across various disciplines in biology. Due to its widespread use in research, daily life, and industry, extensive studies on brewing yeast populations have greatly accelerated humanity's understanding of life.
The Life Cycle of S. cerevisiae
The life cycle of S. cerevisiae has been well documented in the laboratory (Figure 1).
S. cerevisiae usually grows as a diploid in artificial nutrient-rich medium such as YPD (1% yeast extract, 2% peptone, and 2% dextrose) and reproduces clonally by budding, with an optimal growth temperature of around 30◦C. It will sporulate and undergo meiosis in response to nitrogen starvation, resulting in the formation of four haploid spores in an ascus. Two of the spores have mating type a (MATa) and the other two MATα. Mating type is determined by a single locus MAT in the middle of the right arm of chromosome III.
A pair of spores with opposite mating types can mate within the ascus upon germination (intratetrad mating or automixis) and form a diploid cell. Ascospores can also germinate to form haploid cells, which can reproduce mitotically by budding, resulting in the formation of MATa and MATα haploid clones. However, the haploid phase usually only exists for a very short period in the life cycle. A haploid cell can mate with another haploid with an opposite mating type either from a different ascus of the same strain (selfing) or from a different strain (outcrossing or amphimixis). Haploid cells can also undergo a mating-type switch by exchanging types at the MAT locus via a gene conversion event. The molecular mechanism of mating-type switching is the replacement of the genetic factor of the MAT locus by a copy of the alternative factor located at a silent locus. There is one silent locus for each mating type (HML homologous to MATα and HMR homologous to MATa).
Recombination between MATα and HMR(a) or between MATa and HML(α) mediated by the HO gene, which encodes an endonuclease that induces a double strand break of DNA within the MAT locus, results in a switch in mating type. Mating-type switching occurs in an accurately regulated pattern that allows only half of the cells in a colony to switch mating types in any one cell division cycle and produces cells with opposite mating types in close proximity , thus facilitating cell mating to form diploid cells. This process is termed haplo-selfing or autodiploidization. The similar mating-type switch mechanism involving the three-locus (MAT-HML-HMR) factors has been found in the majority of species in the Saccharomycetaceae clade studied. Mating-type switch phenomena mediated by simpler “flip/flop” mechanisms have also been detected in at least 10 other groups of yeasts.
The significance of the life cycle and breeding systems of S. cerevisiae characterized in the laboratory remains largely unknown in natural populations of the species. It is difficult to observe and directly characterize the growth profile, mating behavior and life cycle progress of the speciess in the wild. More ecological and population genetic studies on natural S. cerevisiae are required to examine the roles and consequences of sexual and asexual reproductions of the species in nature.
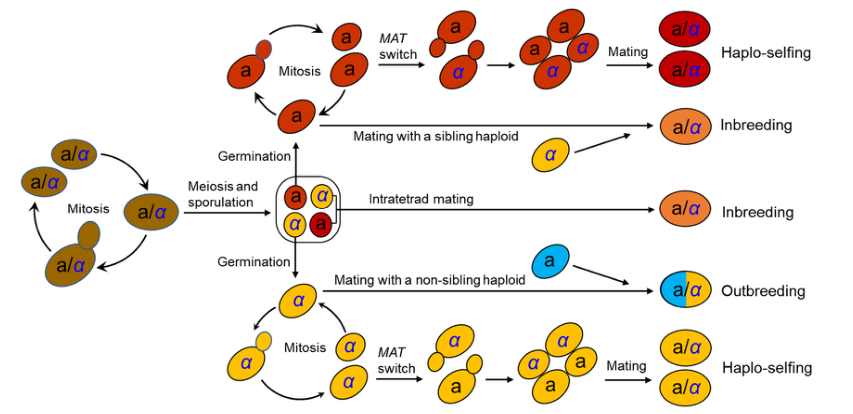
Figure 1 (Bai et al., 2022)
The Ecological distribution of Saccharomyces cerevisiae
For a long time, S. cerevisiae was considered an exclusively domesticated species because of its scarcity in natural environments. Though S. cerevisiae was occasionally isolated from the wild, the feral strains were thought to be the escapees from domestic stocks. However, an early field survey in Japan showed that S. cerevisiae was frequently isolated from forest materials including soil, decayed leaves, and tree bark,implying the common occurrence of the species in nature. Then, a growing number of studies also suggested that S. cerevisiae might be distributed in a wide range of forest habitats as well as vineyards.
The Bai Fengyan laboratory conducted a systematic survey on the distribution of brewing yeast in natural environments. They successfully isolated numerous strains of brewing yeast from samples of fruits, tree bark, soil, and decaying wood, collected from orchards, plantations, secondary forests, and primary forests ranging from tropical to temperate regions. These findings vividly demonstrate the widespread presence of brewing yeast in diverse ecological environments in nature, and for the first time, provide evidence of its extensive occurrence in pristine forests rarely visited by humans.
While it is acknowledged that brewing yeast is widely distributed in natural environments, there are differing views regarding whether this species occupies specific or preferential ecological niches in the wild. Given the dominance of brewing yeast in sugar-rich fermentation environments, it is generally assumed that it should be common in orchards such as vineyards. However, empirical research has shown that the success rate of isolating this yeast from grape berries is actually low. Metagenomic sequencing data also indicate that compared to other yeast groups, brewing yeast is notably rare on mature grapes in vineyards. A field survey reported in 2012 collected 2,064 samples, with results showing that the success rate of isolating brewing yeast(Figure 2) from fruit samples (6.5%) was lower than from decaying wood (9.2%), soil (10.8%), and tree bark (16.5%) samples. The frequency of isolating brewing yeast from forest soil samples (13.7%) was higher than from orchard soil samples (9.1%). Among various fruit samples collected, the success rate of isolating brewing yeast from grape samples was the lowest (1.8%). These unexpected results suggest that grapes or other high-sugar substrates may not be the preferred ecological niche for brewing yeast in the wild. Existing studies suggest that brewing yeast may preferentially inhabit habitats such as Rosaceae plants and certain broad-leaved tree barks, tree trunk secretions, and surrounding soil in the wild, but comprehensive systematic surveys are needed for verification.
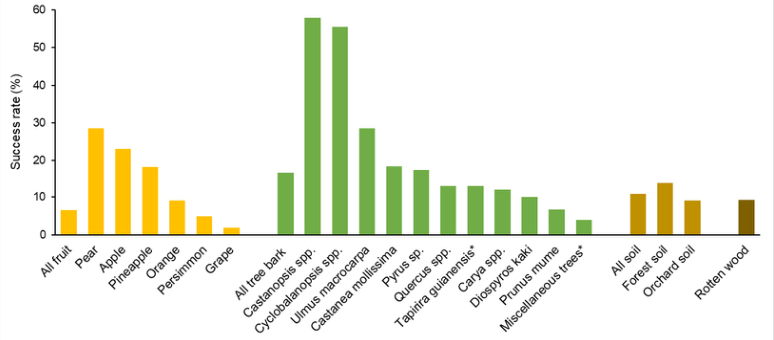
Figure 2 (Bai et al., 2022)
